Here are a few of the current projects going on in the Plough Lab…
Development of an eDNA assay for monitoring and conservation of river herring in Chesapeake Bay
Declining populations of many anadromous fishes on the US east coast has led to increased research on their habitat use and to identify potential impediments to migration (i.e. dams) that are critical to their life history strategy. In the mid-Atlantic region, River herring (comprised of two fishes: Alewife and Blueback herring) abundances have declined over the last few decades and there are renewed efforts to quantify their run numbers and characterize their habitat use across the Chesapeake Bay water shed. Working with Matt Ogburn at Smithsonian Environmental Research Center we are developing an environmental DNA (eDNA) assay to detect and quantify river herring presence across many tributaries of Chesapeake Bay. The basic idea behind ‘eDNA’ sampling is that aquatic species leave behind DNA in their environment (e.g. sloughed cells, gametes, feces), and eDNA from a particular species of interest can be detected from a water sample taken in the proximity of the animal or fish. River herring eDNA can be detected and quantified using a PCR assay that amplifies a targeted, short stretch of DNA specific to river herring – eDNA is ‘extracted’ from small (47mm) filters of surface water samples (1-3 Liters) taken from locations all over the Bay. We are comparing eDNA assay results to traditional ichthyoplankton samples and observations of adult presence absence at the sample locations to test the reliability and sensitivity of the eDNA approach. eDNA data will also contribute to a larger habitat use modeling study led by SERC’s Ogburn.
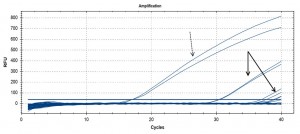
Initial eDNA assay results – image shows raw qPCR output, with PCR cycle number on the x axis. Control samples (fin clip extracted herring DNA) at dashed arrow (early cycles), environmental (water) sample DNA at solid arrows (late cycles).
Striped Bass life history genomics
In common with many other high fecundity marine animals, striped bass (Marone saxatilis) can exhibit substantial fluctuations in yearly recruitment, which is driven by a complex set of environmental factors that affect adult spawning behavior and migration as well as larval survival and production. Understanding the complex interaction of of extrinsic and endogenous (genetic) or behavioral factors and how they shape recruitment variability in Striped bass (and other marine fish) is a major scientific challenge and is of paramount importance for effective management of the fishery. We are using genomic tools to examine the genetic mechanisms underlying variance in spawning behavior (timing, location) migratory behavior (resident/migrant), and larval behavior/mortality in conjunction with the Secor Lab at Chesapeake Biological Laboratory. Currently, we are investigating the possible genetic basis of contingent migratory behavior in adult fish, comparing telemetry data from acoustically tagged individuals in Chesapeake Bay to genome-wide SNP data. As a first step in this work, a high-density genetic linkage map is being created with pair-mated families reared at HPL that will provide genomic position data for future genome-wide association studies.
Cryptic Speciation in Acartia tonsa in Chesapeake Bay
Working with the Pierson lab at Horn Point, we are investigating the genetic and physiological basis of a cryptic species complex of the estuarine copepod Acartia tonsa in Chesapeake Bay. Previous molecular work revealed two cryptic lineages of A. tonsa in Chesapeake Bay structured by salinity (a Fresh and Salt lineage) that are morphologically indistinguishable but appear to be reproductively isolated. This past summer, an REU student in our lab, Adrian Plummer, performed pair crosses within and between the two lineages, revealing hybrid inviability (fertilized eggs produced but almost complete hatching failure) and confirming molecular evidence for long-term reproductive isolation. We are currently working to repeat the crossing experiments and to examine the range of salinity and temperature tolerances of the two lineages.
Population genetics of oyster restoration in Chesapeake Bay
A primary concern with any species restoration or supplementation program is the maintenance of genetic diversity in the restored populations. One way to achieve this is to maximize the number of parents contributing to the gene pool, which should increase the diversity of genotypes that are represented. However, specific features of species life history, practical husbandry constraints, and cost may hinder such efforts. In Chesapeake Bay, oyster restoration programs have focused effort on a few target tributaries that are currently being supplemented via the out-planting of hatchery-produced spat-on shell. These activities have, so far, been successful at increasing biomass locally, and the use of a variety of locally caught wild broodstock has likely kept diversity at a reasonable level. However, high mortality during the early life-stages of oysters and the potential for high variance in reproductive success among parents in mass spawns (i.e. some parental combinations produce more viable offspring than others) may produce cohorts that are represented by far fewer parents than anticipated. For a given cohort of hatchery produced spat-on-shell, if 10 parents spawn initially (producing a mixed parentage cohort), how many of these parents are still represented in the offspring at planting? What is the variance in reproductive success, on average, among parents, and should we be concerned about how many parents are actually contributing to restoration plantings? Does the variance in reproductive success increase at certain stages of production (larval vs spat, or after planting). To address these questions, we are working with the Horn Point Oyster Hatchery to examine genetic diversity at all stages of production. We have performed experiments in which cohorts of larvae produced from group spawns are sampled at various times (early and late larval development,post set,and post planting), and their genotypes compared with parents to determine the relative reproductive contribution among those parents over the course of production.








